Is hereditarianism wrong?
Turkheimer, Gusev, and within-family genetic studies
In the 18th century, French chemist Antoine Lavoisier hypothesized that particles in any matter are held together by an unknown force and pulled apart by heat. The unknown force explains why a piece of iron doesn’t spontaneously explode, while the effect of heat explains why the metal expands and ultimately melts or gasifies over a flame. He thought that heat is a fluid which can flow between the particles and pry them apart.
Lavoisier was more right than wrong. He started with a solid observation – that heat causes objects to expand – and gave a mechanistic explanation of why that happens. He was wrong about the specifics – heat is not a fluid – but he was right about the big picture that objects expand and melt because their particles stay together to a different degree under different conditions. The heat fluid was just a small cog in a greater machinery.
Behavior genetics is one of the very few fields in the social sciences which, just like thermodynamics, has a paradigm, a mechanistic theory about quantifiable elements and testable rules that govern reality. It proposes that the resemblance of family members (a core observation, just like objects expanding over heat) is caused by a combination of genetics and family environment. Behavior genetic studies tend to find a much stronger effect of genetic than environmental forces. Therefore, you can think of hereditarianism as going one step further and saying that it is not the resemblance per se, but the resemblance due to genetic reasons which is the “core observation” in need of further study, such as exploring what biological mechanisms cause family resemblance and to what extent they explain sociological phenomena.
Lately, some researchers, most notably geneticist Sasha Gusev and veteran researcher Eric Turkheimer have been attacking hereditarianism. They are saying that because recent molecular genetic studies were unable to pin down the genetic mechanisms that affect traits, these genetic mechanisms must not exist and the resemblance of family members must be environmental after all. Some of their criticism is well-founded. GWAS has turned out to be a disappointing method, and even after studying millions of people with millions of genetic variants we still cannot explain much about, for instance, hereditary disease.
However, these molecular genetic studies are not a strong test of the basic tenet of hereditarianism, the idea that the resemblance of family members is due to genetic and not environmental forces. They are not analogous to testing whether objects expand over heat: they are analogous of whether there is a “heat fluid”. Just like objects still expand even if there is no heat fluid, genetic forces are still the best candidates for explaining the resemblance of family members even if a very specific idea about what these genetic forces are – the relatively large, linear, additive effects of SNPs picked up by commonly used arrays – turned out to be wrong.
Below is the longer story of why this is.
How we look for genetic effects
There are two kinds of behavior genetics: quantitative and molecular.
It’s easier to do quantitative genetics. Twin studies are like this. You don’t even need real genetic information. Actually, you don’t even need to know what a gene is – people did twin studies starting in the 1920s, three decades before Crick & Watson. In a quantitative genetic study, you study members of the same family – often twins, who have the same age and grew in the same womb which gets rid of important confounders – and check if their phenotypic similarity scales with the degree of relatedness (genetic similarity). The idea is that while both genes and family culture could make members of the same family more similar, we can zero in on just the genes by only looking at pairs of people from within the same family, ideally those living together.
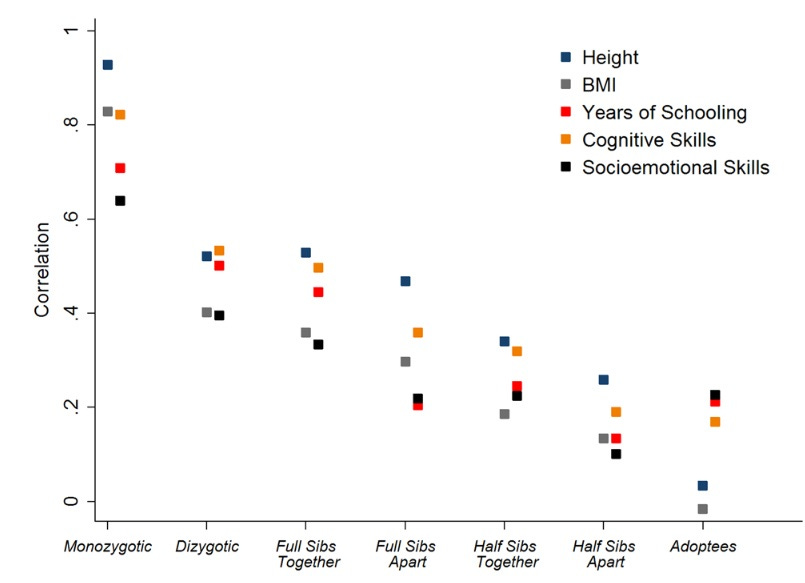
Quantitative genetics at work (source). Pairs of Swedish conscripts from the same families are increasingly more similar if they are more genealogically related. For some traits (height, BMI) this is more true than others (schooling, socioemotional skills), suggesting different degrees of heritability.
They all live in the same house in the same neighborhood, have the same books at home, same income, same parental education, so if half siblings are more similar than adoptees, full siblings than half siblings, MZ twins than full siblings or DZs, then we have evidence that genes matter. By analyzing the degree of similarity – with Falconer’s formula or with fancier path model approaches – we can be more precise and assign a specific number to the relative importance of genetic effects (heritability), of the rearing family which is considered very important by sociologists and psychoanalysts (shared environment), and everything else, including measurement error (nonshared environment). If within-family phenotypic similarity scales perfectly with genetic similarity (MZs are twice as similar as DZs), shared environment is zero. This is often the case. If, additionally, MZ twins are exactly alike, the nonshared environment is also zero and heritability is 100%. This rarely happens, maybe the closest case is height with a heritability over 90%. This logic of quantitative genetics is not flawless, it is not without assumptions (more on whether these hold later), but it is a solid framework to look at the causes of individual differences. The observations in quantitative genetics are the “objects expand over heat” of hereditarianism. You cannot sweep this off the table just because you found a new method.
Molecular genetics relies on knowing the actual DNA sequence. It is newer, much more high-tech, and in a way more serious. Actual genes instead of some vague info about relatedness! Proponents are correct that molecular genetics has the potential of replacing quantitative genetics entirely, because all the information available for the latter – and much more – is theoretically available for the former. But this is true only in theory because you need both the data and a good model of how genetic data leads to traits.
There are basically two ways of using molecular genetics in behavioral research:
- Quantitative genetics with extra frills. We can use the DNA sequence instead of genealogical information to estimate the degree of relatedness between people, and use the quantitative genetic logic from there: are more genetically similar people also phenotypically similar? You do this either with strangers of family members, because everybody is related to everybody else to some degree (do you know your fourth cousins?) and because even family members of the same degree randomly jitter a bit around the degree of genetic similarity expected from genealogy. Using strangers is the way of GREML-GCTA and similar methods, and family members are used in sib-regression and RDR.
- Looking for the genes. This is the real deal: genome-wide association studies (GWAS). This time, we are actually looking for genetic variants causing a trait: the molecular background of intelligence, personality, disease etc. We do this by getting molecular genetic and phenotypic information from very many people and simply looking for genetic variants which are more common in those with a higher level on a trait. Because we have very many genetic variants, we must set the evidentiary bar very high to avoid false positives. When we are done, we can create a formula for a polygenic score: a weighted sum of all the variants we found, a genetic predictor of the trait. We can test in new samples how well this works, we can philosophize about the nature of the variants we found (“height genes are express in the bones, IQ genes are expressed in the brain”) and we can use the score to make predictions about people or groups with known genes but unknown trait levels, such as an embryo.
However, while molecular genetics looks like a more mature method, it still has assumptions. If these are violated, the results can be off – false negatives and false positives are both possible. The assumptions are:
1. Gene effects are additive. One SNP=one unit trait increase, two SNPs: two units. This is reasonable because of what R. A. Fisher realized in his theory of geometric evolution way before DNA was even known. There is no such thing as a “gene for intelligence”: pleiotropy is widespread, genes do many things, if you have a new mutation that raises your IQ you will have other changes as well and they are more likely to be negative than positive. If you want to evolve high intelligence, you are best off with many genetic variants causing this, each with a small effect (so the pleiotropic side-effects cancel out) and each independent of the others (so your descendants are guaranteed to get the IQ raising effect regardless of what their other genes are like).
2. We know all the genes. This is iffier. Most molecular genetic studies see a few hundred thousand common SNPs, specific genetic markers with a lot of variability between people. But there are rare SNPs only a few people have, non-SNP genetic variation (CNVs, indels etc.), and if we go crazy, mitochondrial DNA and maybe some non-DNA mechanism of inheritance (microbiome, epigenetics, some totally undiscovered mechanism?). Both the GCTA and GWAS approaches only see a part of all genetic information: the additive effect of common SNPs. If there are others, there are false negatives. There are also false negatives if we don’t have enough power to detect small effects. Given that even for height four times as many SNPs and almost twice as much variance was found by going from 700k to 5M people, this is not a small issue.
3. There is no stratification. This is the assumption which can result in false positives if violated. Just because a genetic variant is associated with a trait it doesn’t mean it causes it. Asian people usually have a special version of the EDAR gene and can eat with chopsticks – but EDAR is not the “chopsticks gene”, it’s just that there is an accidental overlap between people with the EDAR gene and chopsticks-eating culture.
Ancestral (left) and novel East Asian (right) versions of the EDAR gene. Sorry Gregory Cochran, after your blog’s name I’m also stealing your meme.
A less on the nose (but more realistically problematic) version of stratification is if genetic effects index a shared environmental effect. One version of this is a dynastic effect. Certain genetic variants are probably more common in the Rothschild family, but they don’t make you rich, they just show where you are from, and that does. In a naïve population-wide study, they can still end up being associated with income as proxies of Rothschildness. Another version of genetic nurture, where the genes change parental behavior and parental behavior changes you. You also inherit the genes, but this is not the important part, it was your mother having had them when you were little. Both are cases of passive gene-environment correlation: the genes being stand-ins for environmental causes.
These assumptions are different from those in quantitative studies, so if the two methods result in similar findings, we can be happy that these are robust. But if they do not, we don’t know if the quantitative or the molecular findings were wrong. There is no natural hierarchy between these methods: molecular genetics is not superior to quantitative genetics, it is not the next step in the evolution of the field, merely a new method. They both have strengths and weaknesses. Quantitative genetics uses more complete information so would expect it to see more genetic effects, at the cost of less biological information about the precise nature of them. Molecular genetics is much better for discovering mechanisms at the cost of likely missing the larger picture. You can think of it as looking for a specific kind of “heat fluid”. If it’s not there, thermodynamics is still real, it’s just that you need to find a different proximal mechanism.
If the two methods have matching results, it’s great. If we had a polygenic score that accounted for 80% of intelligence variance, we would have shown that not only is intelligence highly heritable, but caused by common SNPs with a linear effect large enough to be seen in our current sample. The fact that we don’t simply doesn’t say anything about the heritability of intelligence, merely that a very lucky outcome didn’t come to be.
The GWAS embarrassment
Modern molecular genetics turned out to be something of an embarrassment. In the early 21st century people believed that just knowing what genes there are and how they work – the Human Genome Project – would be enough to understand all apparently hereditary disease. For example, Francis Collins, director of the Human Genome Project said in a 2000 interview that:
“In the next five to seven years, we should identify the genetic susceptibility factors for virtually all common diseases — cancer, diabetes, heart disease, the major mental illnesses — on down that list. Out of that will come the ability to make predictions about who’s at risk and, in many instances, that itself can be extremely helpful by allowing you to take advantage of surveillance, so that you are diagnosed early before a real problem arises. In the longer term — perhaps ten or 15 years — this will lead to a new generation of therapies that is going to be much more successful and effectively targeted towards the basic problem than what we have now.”
, and the next year confirmed this opinion in a testimony in front of the US Senate.
Needless to say, no such thing has happened. Forget about intelligence and other based stuff for now: molecular genetics just doesn’t work well, even in medicine. In a typical modern GWAS, researchers look at data from hundreds of thousands, sometimes millions of people to figure out which genetic variants – or a combination of them, a polygenic risk score – are associated with certain traits. You would expect that with so much information and such high tech approaches – SNP arrays which digitize molecules to a TXT file, arcane statistical approaches, the known biological function of thousands of genes – they would be able to learn a lot. But they don’t.
I looked at some recent medical GWASes which looked state of the art, and they were unimpressive. In this study, using genetic information from 13790 bladder cancer cases and 343502 controls the researchers could construct a genetic score which predicted about 50% of extra cancer risk per standard deviation. There are similar figures for heart disease. A trick I often see in these studies is that the authors focus on “new loci”, specific genetic variants associated with the trait. Their discovery is claimed to be a big deal, and they write lengthy paragraphs with a lot of biology and chemistry about what the genes close to these variants may do to cause the disease. This makes the paper science-y and keeps the grant money flowing. But each of those variants confers maybe 0.001% extra risk! All these speculations about mechanisms are an exercise in futility because the causal link from gene to trait is just not there. The observation that we cannot really find the genetic variants underlying the heritability suggested by quantitative genetic studies is called the missing heritability problem.
Based traits like educational attainment or intelligence are actually among those with better GWAS results. The most recent educational attainment GWAS, based on over 3 million people, could build a genetic predictor which correlated about ~0.37 with actual educational attainment, which is not bad given that the number we are trying to replicate is ~0.65 (the square root of a meta-analytic twin heritability of 0.43). This predictor also had a decent correlation with cognitive ability. Not bad, but 20 years ago we would have expected much better with 3 million genomes.
Also, for these traits, there is a particular embarrassment. In 2018, a group of Icelandic researchers discovered that your parents’ genetic variants explained about half as much variance of educational attainment than your own, even if you did not inherit these. This led to the recognition of indirect genetic effects: the idea that genetic variants can be proxies for environments instead of true biological causes. These are the “genetic nurture” and “dynastic” effects I wrote about, and they mean that even of the small effect of discovered genes a big part is an illusion. To address this, modern studies use within-family designs: for example, in a GWAS they would compare sibling pairs, not a group of random people, to discover which genetic variants truly cause traits. They usually keep finding these indirect effects, which make GWAS findings even less impressive and missing heritability an even bigger problem. This suggests that, ironically, genetic variants may pick up the admittedly non-zero shared environmental effects on socially important traits better than the actual genetic ones. (Keep in mind that within-family studies are always less powered so they are biased towards null findings.)
Apples to oranges
Turkheimer’s and Gusev’s whole shtick is acting as if molecular genetic studies are a test of “heat expansion” and not just a specific “heat fluid” – that if we are not finding all the genetic effects using the current molecular genetic methods, they must not be there. They play switcheroo with terms, jumping between “heritability” meaning heritability by quantitative methods or how much variance molecular methods explain.
For example, in his debut piece “No, intelligence is not like height”, Gusev claims that “IQ is much less heritable” than height, based on molecular genetic studies with vastly different statistical power to detect genetic effects on the two traits. (See Joseph Bronski’s response to that particular post.)
Turkheimer makes me scratch my head even more, claiming things like that:
“[Hans Eysenck] estimated that the heritability of neuroticism was .81. […] This week, a preprint describing a massive meta-analysis of sibling GWAS dropped. The estimate of the direct genetic effect heritability of neuroticism was .081, down from Eysenck’s estimate by an order of magnitude.”
, confounding a twin result with an underpowered within-family GCTA-style study. (David Hugh-Jones calls him out on this in comments on this piece.) They especially like to pile on the fact that within-family estimates are lower than population estimates, hinting that this means that shared environmental effects are very important and heritability is overstated. But this is a minor thing: the majority of quantitative heritability is not found even in simple between-family molecular genetic studies. Compared to this, the fact that a third of the remaining effect is also gone once we look within families is just salt on a headshot.
Gusev and Turkheimer treat quantitative genetic studies as outdated, messy and based on arbitrary methodological choices. Turkheimer hardly engages with them anymore, semi-joking that a still very small within-family molecular genetic study is “the end of social science genomics” and that not taking molecular genetic results as the true estimates of heritability would be a “Plomin-style” ignoring of “null results”. Gusev tries to be more serious and actually engages with quantitative genetic studies in two posts. In the first, he keeps making the argument that if a molecular method (RDR) finds lower heritability estimates then the quantitative estimate then the latter is wrong, which, as we saw, is a fallacy. In the second, he writes that “twin heritability models can tell you whatever you want to hear” and dazzles the reader with complicated equations and path models used in twin modelling. He singles out one galaxy brain paper, Bingley et al 2023, as evidence that heritability can be made very low and shared environmental influence very high. This paper gets this result by assuming equal avuncular shared environments, in other words, by making the clearly wrong assumption that all differences between the families of cousins are environmental. [Sasha Gusev and Unboxing Politics both pointed out in the comments that this is a wrong reading of the paper’s assumption. I’ll have to get back to this paper - let it be known that everything I write is potentially wrong and I acknowledge the errors I make.]
At this point, let’s circle back to the original analogy, that our undeniable core observation is that “family members are similar”, much like how Lavoisier’s observation was that heated objects expand. Lots of different things could be pushing heated molecules apart, so falsifying the heat fluid theory still leaves the chemist with a lot of different ideas to explore. However, by definition there are only two things that can make family members similar: genes or the shared environment. By denouncing genetic effects, Gusev and Turkheimer are implicitly saying that shared environmental effects account for almost all of the similarity of family members.
Now I’m not a bigoted hereditarian and I wrote a piece about the importance of the shared environment myself, but this is a very tall order. Because of quantitative genetic findings – the only kind which can find shared environmental effects – it is extremely unlikely that shared environmental effects predominate.
The missing shared environment
Sasha Gusev is right that quantitative genetic studies have their own assumptions. But in some designs, the assumptions are minimal. Plus, different designs have different assumptions so it would be a big coincidence if they all produced the same fallacious result.
One design is monozygotic twins reared apart (MZAs). These are people with the same genes raised by different families: a perfect test of genetic effects without shared environmental confounding. MZA studies are very hard to do and there are roughly two samples with a reasonable size: the America MISTRA and the Swedish SATSA. Both concur that, for example for IQ, being reared together doesn’t matter much. (The MISTRA paper discusses lots of other phenotypes as well.) I made this mashup of results from the SATSA paper and this one:
On the upper left you can see the intelligence twin correlations in SATSA. Note that MZAs are just as similar as MZTs. Hello, Jay Joseph, this has DZA data too! In MISTRA, the MZA-based heritability estimate was 0.81 so almost exactly the same. One could quibble (see Kaplan 2012) that 1) MZAs were not raised completely apart, they had some contact, and 2) members of the same pair were adopted by similar families (selective placement) so in a way they did have some shared environments. On the chart I’m showing on the right, Bouchard et al show how 1) is wrong: there is no relationship between contact duration and similarity (and also note that MZA pairs were not much less similar than the same person tested twice!). The table on the bottom shows that 2) is wrong too. Bouchard et al show that selective placement is real (socioeconomic characteristics of the adoptive families are correlated) but controlling for it doesn’t change the twin correlations. Simply put, MZAs ending up in more similar families don’t end up more similar. If identical twins have similar IQs because their parents raised them a certain way – as implicitly claimed by Turkheimer and Gusev – then why does the shared environment appear so inconsequential?
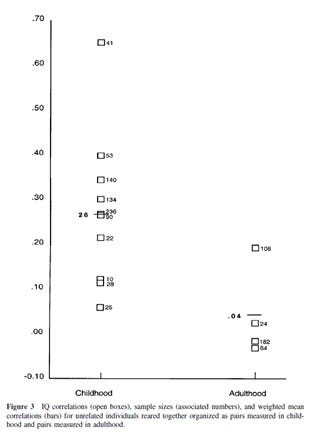
We can of course look at the shared environment directly by studying adoptees. Do children growing up in the same family end up similar? Yes, as long as they actually live in that family. Bouchard & McGue’s classic chart about adoptee IQ correlations:
There are only a few studies which were published since then, some in Nancy Segal’s virtual twin samples (similarly aged adoptees) and this one in adults. These replicate Bouchard’s initial results very well. The virtual twins have somewhat similar IQs (r~0.25), but this is unsurprising: they were still children when they were studies and families do influence the IQs of small children. But in the adult study this is only at 0.06, not even statistically significant. For the sake of simplicity I focused on IQ, but see my previous post about attachment and Inquisitive Bird’s great shared environment post for other traits. If growing up in a certain family is so important and genetic effects are an illusion, why do adoptees end up barely or not at all similar?
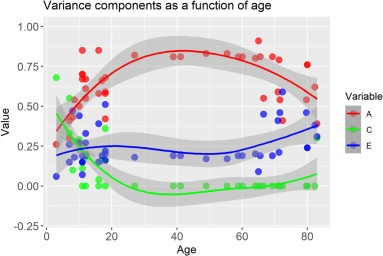
Next up: twin studies. Gusev releases a lot of intellectual squid ink when he dazzles you with the many ways one can analyze twin data: in truth, most twin studies are fairly simple, they just assume additive genetic, shared environmental, and nonshared environmental effects. Basically, they assume that twice the excess similarity of MZs over DZs is heritability. These studies tend to show high heritability and low shared environmentality in basically whatever trait you use them for, something Turkheimer himself famously noticed 25 years ago. But there is a twist: this depends on age. In children, heritability tends to be lower and the shared environment is still important, whereas adults grow out of whatever their parents did to them and regress to their genetic tendencies, so heritability grows. This is known as the Wilson effect, which shows up for intelligence, psychiatric problems, attitudes, body mass index, and even attachment.
Note that this is exactly the same we saw in the adoption literature.
Let’s assume that twin studies are done wrong, it’s not the genes but the parents that matter, MZs have uniquely similar parenting experiences, and this, not the genes, makes them more similar than DZs. Why is this most evident in adults who are long done being raised by their parents, one way or another? Why isn’t this mirage heritability strongest when parents are the most active, and why don’t children from the same family regress back to the relatedness-independent effects of the parenting they experienced, erasing MZ-DZ differences? The Wilson effect is evidence that the twin method can very much produce high shared environmental effects and low heritability: it totally does this for children, just not for adults. A hereditarian can easily explain this with Warne’s first law of behavioral interventions: the shared environment’s effect fades when its gone, but genes stay with us lifelong so ultimately they get the upper hand. An anti-hereditarian cannot.
We can also test some twin study assumptions directly. Quantitative genetic studies check if the similarity of pairs of family members scales with genetic similarity. Normally, there are two things that can make family members similar: family culture (shared environment) and genetics. By only studying pairs of people from within the same family, we are holding the first constant and we are only looking at genetics. If a trait is heritable, we expect full siblings to be more similar than half siblings and MZ twins more similar than either DZs or siblings. Twin studies are the best kind of quantitative study in many ways because twins are the same age and gestated in the same womb, so we don’t have to worry about these confounders.
An assumption of this method is that family culture is distributed evenly and genetically more similar family members don’t, for example, get more similar parenting. This is called the equal environment assumption. As it happens, this is wrong. MZ twins are reared more similarly: for example, parents often dress up MZ twin kids in the same clothes, but this rarely happens for DZs.
Felson (2014) is perhaps the nicest illustration of why the equal environments assumption doesn’t matter. MZs really have (and had, as children) more similar environments – we know this from self-report scales (in this study, from the American MIDUS sample). But these self-reports can be used as covariates in twin analyses: regardless of zygosity, do twin pairs with more similar environments turn out more similar? Felson says no:
We see here the twin heritability of many traits, measured in the ordinary way (circles) and after controlling for environmental similarity, which gets rid of the problem of unequal environments (at least as long as the 15 measures of these capture whatever is relevant). The heritabilities barely move: the only statistically significant drop is for neuroticism, which becomes slightly less heritable after adjusting for environmental similarity[1].
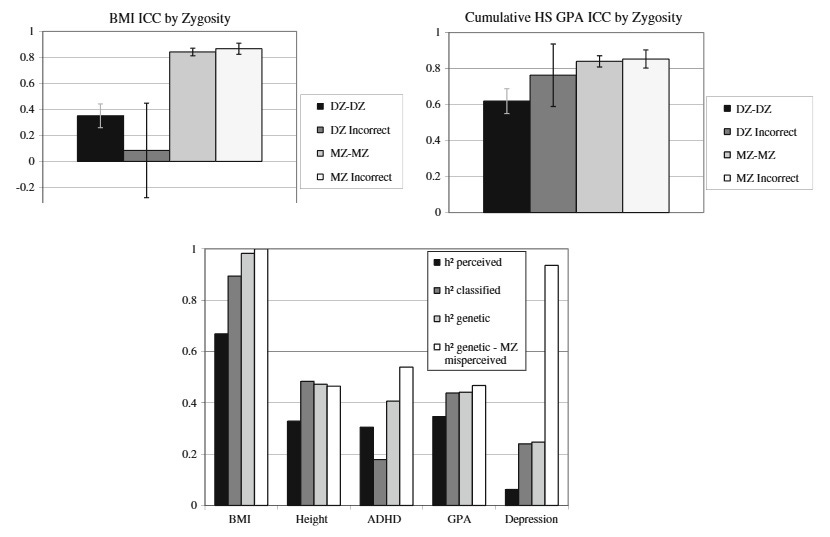
An even more straightforward way to test the equal environments assumption is to use misclassified twins. As it turns out, parents of twins are frequently wrong about the zygosity of their kids. Misclassified twins – MZs believed to be DZs or vice versa – should get a mismatched genetic and environmental package so looking at the degree of their similarity is a good way to gauge which one matters more. If the degree of genetic similarity is the key, misclassified twins should be as similar as their genes predict, and if parenting matters more, they should match the “social zygosity” their parents assign to them. The most comprehensive study I know about this is Conley et al 2013, who checked the parents’ beliefs about zygosity using genetic information and found that genes matter much more:
I parsed together three of their figures. The top two show the similarity of misclassified twins (DZ incorrect: DZs raised as MZs, MZ incorrect: MZs raised as DZs) compared to those correctly classified. It is much more common for MZs to be raised as DZs (almost a quarter of all pairs!) and they are just as similar as ordinary MZs. DZs raised as MZs, on the other hand, don’t catch up to real MZs in terms of similarity, but it’s hard to say anything precise about this due to low sample sizes (12-16 pairs). On the bottom chart, the authors show that this misclassification problem is not a trivial problem in twin research: Heritability is lower if you go by parental reports of zygosity (“perceived”), or even a standardized questionnaire (“classified”) and higher if you use real genetic information (“genetic”). Not only are equal environments not an issue, true heritability is even higher than seen in most studies which don’t have genetic data!
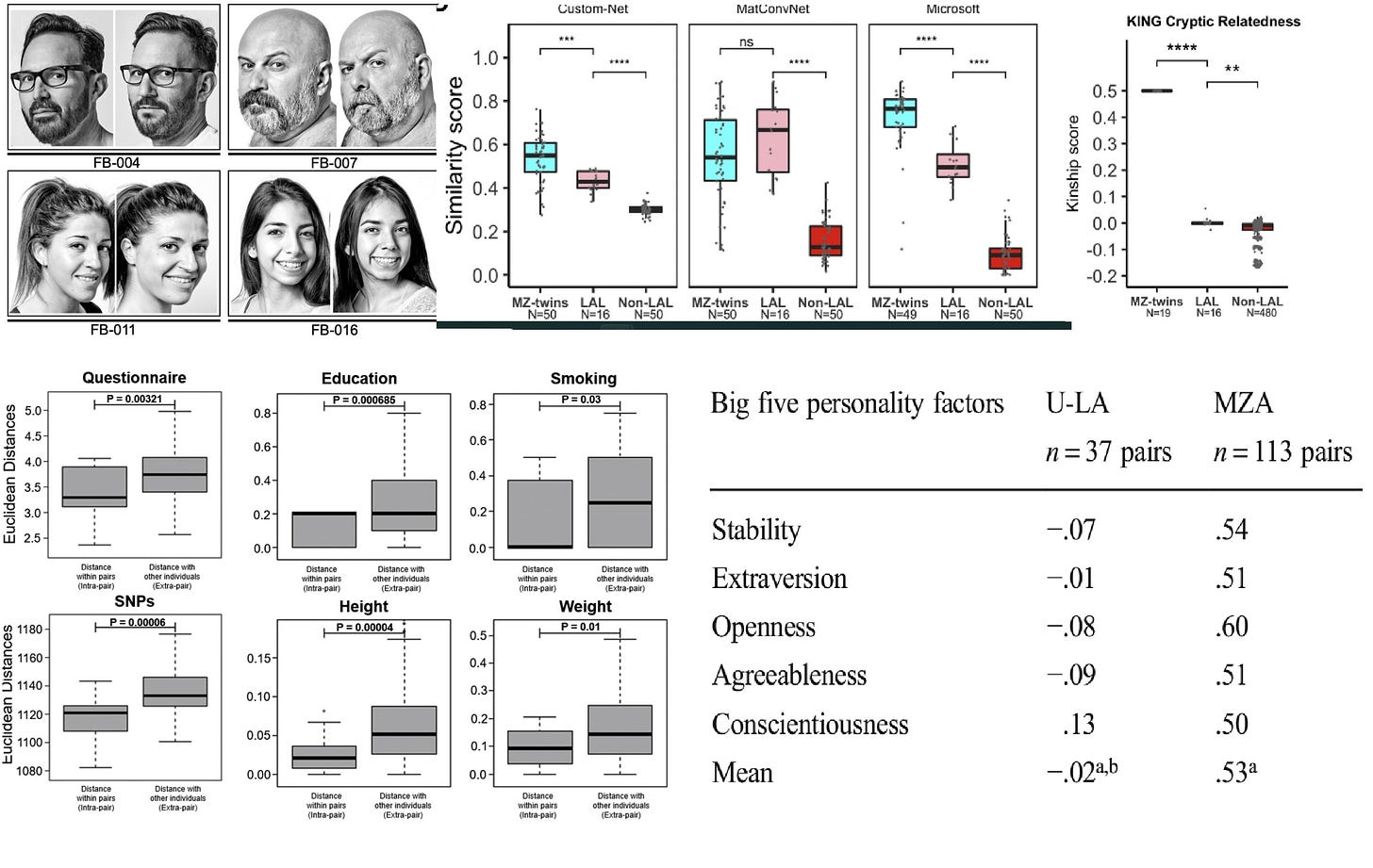
In one comment I found very interesting, Sasha Gusev wonders “how parents (and society) treat kids when they look identical”. This is important for us because maybe MZs are so similar because other people – not just your parents – react to their appearance in a certain way. It’s harder to make it in life if you are an ugly person. As it happens, there is direct research about lookalikes, people who look very similar despite being strangers: I’m aware of at least four such studies.
To me, lookalikes look like MZ twins (top left) but based on three algorithms they are actually between random people (non-LAL) and MZ twins and they are slightly, but significantly more genetically related than random people (top right). The Cell Reports paper I linked found that these look-alikes are slightly more similar in some traits – mainly genetics, height, and education – than random pairs of people (bottom left), but unfortunately didn’t directly compare them to twins. This was done in all the other linked studies by Nancy Segal, however. One example is shown on the bottom right: MZ twins, as we know, all have substantial personality similarity but lookalikes do not. There is not much evidence that how parents and society treats us when we look a certain way has a strong influence on us: it certainly isn’t the main reason MZs are so similar.
You might say that hereditarianism is wrong, but you cannot deny that family members are similar. If you don’t think this is genetic, you are saying it is shared environmental: there is nothing else that makes family members similar. I fail to see how the results with MZAs, adoptees, the Wilson effect, misclassified twins, or lookalikes are compatible with genetic effects being an illusion and the shared environment being dominant.
[1] To be fair, the trend is downward in all the other cases, but I’m autistic enough to notice that this method overcontrols for environmental similarity. This is because the environmental similarity measures might themselves be heritable and genetically correlated with the traits, which means that by controlling them we are getting rid of genuine genetic effects. Maybe saying “my twin cares about me” is not a totally objective measure of what this question means, but taps into a heritable tendency to trust others which is also captured by the neuroticism test. In this case, controlling for the former gets rid of some of the bona fide genetic effects on neuroticism, not just the effect of really having a caring twin.




It is unfortunate that while this post makes more of an effort to engage with modern data than the typical response, it also makes several egregious errors that ultimately undermine that effort.
1. "Gusev claims that “IQ is much less heritable” than height, based on molecular genetic studies with vastly different statistical power to detect genetic effects on the two traits"
This is incorrect. The estimation of molecular heritability does not depend on the power to detect genetic effects at all, it is a single genome-wide parameter aggregated over all variants in the study regardless of whether they are significant.
I explained this point in my first post on IQ with a reference (https://theinfinitesimal.substack.com/p/no-intelligence-is-not-like-height): "But prediction accuracy depends on sample size, could the findings drastically change with more samples in the future? In fact, through the magic of statistics, we actually know that this claim will always to be true. We know this because we have estimated a parameter called molecular heritability, which tells us the upper bound on what a genetic predictor could ever achieve".
And then I explained it again in my second post responding to comments (https://theinfinitesimal.substack.com/i/148251755/isnt-gwas-heritability-only-quantifying-the-mechanisms-we-currently-understand): "Isn’t GWAS heritability only quantifying the mechanisms we currently understand? This is a typical misconception: that GWAS only quantifies the heritability from individual significant associations or genes we understand. In fact, GWAS heritability is defined as the phenotypic variance explained by all genetic variation that has been measured, whether it is significant or not.".
I can explain it a third time, I guess. The population-scale molecular heritability of IQ is 10-20% and the population-scale molecular heritability of height is 40-50%. These estimates come with very low uncertainty, and the fact that 40% is larger than 20% is well established. The inability of this post to grapple with the basic operation of these methods nor their basic estimates should raise red flags about the other claims.
2. "He singles out one galaxy brain paper, Bingley et al 2023, as evidence that heritability can be made very low and shared environmental influence very high. This paper gets this result by assuming equal avuncular shared environments, in other words, by making the clearly wrong assumption that all differences between the families of cousins are environmental."
This is incorrect. Bingley et al. propose a quantitative method to directly test the equal environment assumption, something the author should be pleased to see. Their method assumes equal shared environments between twins and their nieces/nephews as between twin spouses and their nieces/nephews ("To avoid adding parameters, we assume that the degree of environmental sharing between twins and their niblings (Equation 15) is the same as that between twins’ spouses and niblings (Equation 16)"). This does not imply that "all differences between the families of cousins are environmental", it is a constraint *specifically* on the shared environments between these relationship classes. This constraint is clearly much less restrictive than the assumption from classic twin models that MZs and DZs have identical shared environments, which we know they do not. In addition to the Bingley paper, I describe a number of other studies that evaluated the EEA assumption with other extended family designs and found it to be violated (https://theinfinitesimal.substack.com/p/twin-heritability-models-can-tell). Again the author appears not to understand the model but it is confident that the model is wrong because it produces uncomfortable results.
3. "In the first, [Gusev] keeps making the argument that if a molecular method (RDR) finds lower heritability estimates then the quantitative estimate then the latter is wrong, which, as we saw, is a fallacy."
The author first presents RDR as "quantitative genetics with extra frills" but then later discards RDR because it is a molecular method. Well, which is it? What the fallacy is in reporting the RDR results is also never stated. It is odd to simply discount the strongest evidence against one's position as "a fallacy", typically this is the evidence one would addresses first and in most detail. I'll remind readers that the paper that introduced the RDR method is titled "Relatedness disequilibrium regression estimates heritability without environmental bias" (https://pubmed.ncbi.nlm.nih.gov/30104764/) and the entire motivation of the method was to estimate complete additive heritability (using Identical By Descent relatedness estimates) without the environmental assumptions of classic twin models. When applied to a large number of traits, the average RDR heritability was 32%, compared to an average twin heritability of 61% (and a SNP-based heritability of 26%); results that I also summarized in my post, which also goes into how RDR works (https://theinfinitesimal.substack.com/i/148251755/okay-but-why-do-you-think-twin-estimates-so-much-higher). Since RDR and twin estimates are biased to the same extent by assortative mating, this gap is explained by environmental bias in twin studies. So the quantitative genetics (with extra frills) models the author likes also tell us that twin estimates are 200% inflated on average, and that SNP/GWAS estimates are about 80% of the total heritability on average. These relative estimates are consistent with a variety of other studies and theory showing that common variants (i.e. those in GWAS) explain the majority of total heritability, so everything hangs together nicely.
---
TLDR: This post misunderstands how molecular genetic studies actually estimate heritability, then misunderstands how quantitative extended twin models have probed the equal environment assumption and found it violated, then finally does a sleight of hand by pretending that RDR is not a quantitative genetics method because the findings are inconvenient.
Finally, I do appreciate that this post finally acknowledges that behavioral GWAS were greatly inflated by population stratification and, for hereditarians, a disappointment. But I don't think we can simply let hereditarians off the hook, because they were the ones who bet hard on GWAS methods to begin with. Charles Murray predicted for over a decade that molecular studies were just about to prove him right (https://x.com/evopsychgoogle/status/1188441459860443137); Razib Khan argued that we just need to wait a little bit longer for GWAS to fully explain the genetics of intelligence (https://undark.org/2017/02/28/race-science-razib-khan-racism/); Steve Hsu predicted that GWAS would explain 60% of the total variance of intelligence by 2016 (https://x.com/SashaGusevPosts/status/1613724821480677378); Emil Kirkegaard has been running bogus polygenic score comparisons pretending stratification is a non-issue and getting the results completely backwards (https://theinfinitesimal.substack.com/p/how-population-stratification-led). Even setting aside motivated hereditarians, here's what internet celebrity Gwern said about the first GWAS of educational attainment: "Reading it was a revelation. The debate was over: behavioral genetics was right, and the critics were wrong. Kamin, Gould, Lewontin, Shalizi, the whole sorry pack—annihilated. IQ was indeed highly heritable, polygenic, and GWASes would only get better for it" (https://www.lesswrong.com/posts/sRchPdp6mCqY2ekJX/what-progress-gwern-s-10-year-retrospective). As we now know, this is not what actually happened.
And this ultimately gets at my issue with the modern hereditarian movement, my point is not to attack their *theories*, everyone is entitled to speculate about genetic processes and test out their speculation. What I am attacking is the pattern of overfitting and p-hacking in their *methods*. When GWAS provided a cudgel to beat up on Lewontin and Shalizi, then suddenly it was "a revelation" that will only get better and better. Now that GWAS falsifies these claims, it is deemed "something of an embarrassment". When the RDR method is first described, it is just "quantitative genetics with extra frills" (laudatory) but when it yields uncomfortable results, it is now "molecular genetics" (derogatory) and using it is "a fallacy"". Methods and findings are not used to understand how the world works, but to stitch together a narrative that supports the author's predetermined conclusion.
Nice post. But I'm a bit bummed because I have been writing a similar piece for some time...
I guess I have to make it better now.